Translational Control
The ribosome has been a driver for single-particle cryo-EM methods development since the 1970s. We have a long-standing collaboration with the Korostelev lab (University of Massachusetts Medical School) on translational control of gene expression by the ribosome. In a recent example, we visualized the stepwise decoding of cognate aminoacyl-tRNA [1] [2]. Using the “focused mask” feature of Frealign, we were able to resolve small structural differences between three states of tRNA accommodation in the A site of the small subunit of a bacterial ribosome. The accommodation of cognate tRNA, and rejection of other tRNAs is a fundamental requirement for fidelity in ribosomal translation. Our work suggests that the 30S subunit only efficiently closes when a cognate tRNA is bound, moving the elongation factor EF-Tu into position to be activated for GTP hydrolysis by the sarcin-ricin loop.
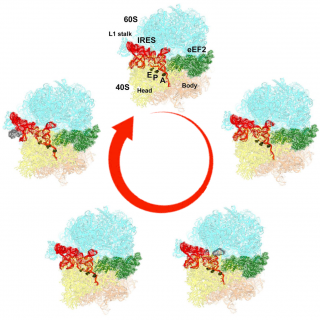
Furthermore, with the computational efficiency of Frealign, we were able to resolve 10 different states of a eukaryotic ribosome prepared with elongation factor eEF2 and IRES (internal ribosome entry site) RNA (Figure), an mRNA from Taura syndrome virus (TSV) that initiates translation in a non-canonical fashion [3]. The visualization of so many states present in a single sample (termed "ensemble cryo-EM") requires large datasets, typically over one million particle images. In the case of the IRES RNA, our work showed that TSV IRES is translocated from the A site to the P and E sites in an “inchworm-like” manner by inter-subunit rotation (ratcheting) and eEF2 moving into the A site to block reverse translocation.